Developing a Linear Regression Algorithm
The Linear Regression is specified as;
where $X_0 = 1$
and;
$(\theta_0, \theta_1, \theta_2,…, \theta_n)$ are the optimized thetas,
$(X_1, X_2,…, X_n)$ are the features or independent variables.
where our algorithm learns from our training data and outputs a prediction hypothesis $h_{(\theta)}(X)$ function which is used to predict actual values of $Y$.
where $\epsilon$ is the Error term.
There are two ways to compute these optimized parameters;
- Gradient Descent
- Normal Equation
A general overview of the process;
- Import required libraries
- Develop model using both Gradient Descent and Normal Equation
- Test Model using both methods on a dataset and compare with Sklearn out-of-the box model using the Root Mean Square Error (RMSE)
Let’s import the important libraries:
import numpy as np
import pandas as pd
from matplotlib import pyplot as plt
from sklearn import datasets
from sklearn.model_selection import train_test_split
from sklearn.metrics import mean_squared_error
1. Gradient Descent
m = Number of Samples
def cost_function(X, y, init0s): #Returns the cost value given the theta values.
m = len(X) #m is the length of X i.e the number of samples
cost = 1/(2*m)*np.sum((np.dot(init0s, np.transpose(X)) - y)**2)
return cost
def partial_derivs(X, y, init0s): #Returns a list of updated thetas
m = len(X)
predicted = (np.transpose(init0s) * X).sum(axis = 1)
diff = (predicted - y)
diff_transpose = np.transpose(diff[:,np.newaxis])
#Add a new axis to make it vectorizable for dot product
updated0s = init0s - alpha*((1/m))* np.dot(diff_transpose, X)[0,:]
#Remove the extra dimension/axis
return updated0s
def gradient_descent(X, y,init0s):
#Returns the optimal theta value and a list of costs for each iteration
costs = [cost_function(X, y,init0s)]
for i in range(epochs):
init0s = partial_derivs(X, y,init0s)
c = cost_function(X, y, init0s)
costs.append(c)
return init0s, costs
def predict(X_test, y_test, optimal0s):
predicted = (np.transpose(optimal0s) * X_test).sum(axis = 1)
rmse = mean_squared_error(y_test, predicted)
return predicted, rmse
Let’s test out our Linear Regression model on a dataset
data = datasets.make_regression(100,2, noise = 1) #100 samples with two features
print(data)
(array([[ 0.0465673 , 0.80186103],
[-2.02220122, 0.31563495],
[-0.38405435, -0.3224172 ],
[-1.31228341, 0.35054598],
[-0.88762896, -0.19183555],
[-1.61577235, -0.03869551],
[-2.43483776, -0.31011677],
[ 2.10025514, 0.19091548],
[-0.50446586, -1.44411381],
...
[-2.06014071, 1.46210794],
[-0.63699565, 0.05080775],
[ 1.25286816, -0.75439794],
[ 1.04444209, 0.81095167],
[ 1.23616403, -1.85798186],
[ 2.18697965, 1.0388246 ],
[-0.07557171, 0.48851815],
[ 0.59357852, 0.40349164]]), array([ 72.36523412, -37.14156866, -41.38412368, -12.64791154,
-43.66708325, -55.37682786, -104.14429178, 86.72486169,
-141.78457722, -57.09382215, 21.11295519, 37.8001059 ,
199.5500905 , 25.81364514, 209.42911479, 128.20604958,
-26.3460515 , -88.2946975 , 45.84342752, -29.55352689,
...
102.81595456, -28.66364244, 100.21268739, -73.03008828,
113.01648102, 109.17545836, -31.30447011, 36.51499677,
-88.82089559, -3.4467088 , 53.45209032, -87.40808263,
-99.40016109, 5.04506704, 56.12030581, 84.54855562,
59.02884225, -17.38215753, -24.1274554 , 102.33610869,
-120.59599797, 160.11974736, 39.67236875, 55.97220222]))
We need to carry out some preprocessing on data.
- Add the $X_0$ column with 1’s
- Split the data into Train and Split for Validation
def preprocess_data(data):
X = data[0]
y = data[1]
#copy = data.copy
ones = np.ones(len(X))
X = np.c_[ones, X]
return train_test_split(X, y, test_size=0.33, random_state=1)
X_train, X_test, y_train, y_test = preprocess_data(data)
#y_train
Plot the Cost values
epochs = 1000 #Number of Iterations
alpha = 0.04
init0s = np.zeros(3)
y = gradient_descent(X_train, y_train, init0s)[1]
x = np.arange(len(y))
plt.xlabel("Number of Epochs")
plt.ylabel("Cost")
plt.plot(x,y)
[<matplotlib.lines.Line2D at 0x7f2d50903ac8>]
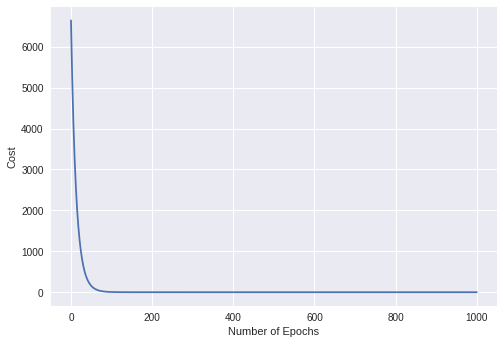
Optimal Theta Values
gradient_descent(X_train, y_train, init0s)[0]
array([ 0.19357013, 32.19125678, 86.08455637])
So our multivariable linear regression prediction model is;
$h_{(\theta)}(X)$ = 0.19357013$X_0$ + 32.19125678$X_1$ + 86.08455637$X_2$
Compare our model’s RMSE with Sklearn’s
epochs = 1000 #Number of Iterations
alpha = 0.04
init0s = np.zeros(3)
predict(X_test, y_test, gradient_descent(X_train, y_train, init0s)[0])[1]
#Returns the model RMSE
1.4033255546659869
from sklearn.linear_model import LinearRegression
lr = LinearRegression()
lr.fit(X_train, y_train)
prediction = lr.predict(X_test)
rmse = mean_squared_error(y_test, prediction)
rmse
1.4033255545376973
Our model’s RMSE value (1.40332) is comparable to Sklearn’s RMSE value (1.40332)
2. Normal Equation
where $\theta_j = (X^TX)^{-1}X^TY$
def normal_equation(X, y):
xtrans = np.transpose(X)
inv = np.linalg.pinv(np.dot(xtrans, X))
#use pinv for nonivertible singular matrices and use inv for normal invertible matrices
optimal0s = np.dot(np.dot(inv, xtrans), y)
return optimal0s
normal_equation(X_train, y_train)
array([ 0.19357013, 32.19125678, 86.08455637])